Projects
We are a multidisciplinary research group working at the intersection of biophysical chemistry, mathematical modeling, and pathophysiology. We believe that there is a continuum between health and disease. If we are capable of measuring this continuum, we can revolutionize early disease detection, deepen our understanding of underlying mechanisms, and develop more precise interventions.
Through our diverse research projects, we leverage our interdisciplinary perspective to provide innovative solutions to uncover new insights into disease mechanism, develop cutting-edge diagnostic tools, and design more effective therapeutic strategies.
Principles of Enzyme Kinetics
We develop standard-based approaches for measuring enzyme kinetic parameters and deriving rate equations that distinguish the mechanisms of enzyme-catalyzed reactions
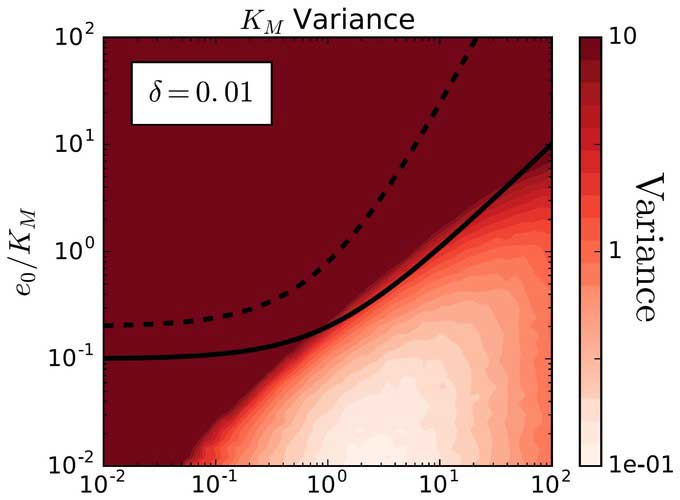
We combine chemical kinetics with mathematical, computational, and statistical methods to develop standard-based approaches, which can distinguish molecular mechanisms under various experimental conditions.
Our research focuses on deriving mathematical approximations of rate law equations that govern the progress curves of enzyme-catalyzed reactions. This involves implementing advanced mathematical techniques for scaling and perturbation analysis. After deriving these approximations, we assess their effectiveness in modeling progress curves and estimating kinetic parameters under different experimental conditions. In addition, we develop and implement algorithms to accurately estimate enzyme kinetic parameters directly from progress curve experiments.
Our work in enzyme kinetics has broad implications for understanding the fundamental principles of biochemical reactions and can inform the development of clinica test, novel therapeutics and biotechnological applications.
Developing Standards for Reporting Enzymology Data
We are developing standards for data reporting enzyme functional data with the aim to improve the quality of data published in the scientific literature.

We are developing Standards for REporting ENzymology DAta (STRENDA) in collaboration with an international committee sponsored by the Beilstein-Institut.
The STRENDA Guidelines are developed through extensive interactions with the biochemistry community to define the minimum information that is needed to correctly describe assay conditions (List Level 1A) and enzyme activity data (List Level 1B). STRENDA aims to ensure that data sets are complete and validated, allowing scientists to review, reuse and verify them. The emphasis is on providing useful and reliable information.
STRENDA Dabatabase
Standards for REporting ENzymology DAta DataBase (STRENDA DB) is a validation and storage system for enzyme function data that incorporates the STRENDA Guidelines. It provides authors who are preparing a manuscript with a user-friendly, web-based service that checks automatically enzymology data sets entered in the submission form that they are complete and valid before they are submitted as part of a publication to a journal.
EnzymeML: An Open Standard for Sharing and Standardizing Enzyme Kinetics Data
The EnzymeML language is an open standard based on the XML markup language.
EnzymeML is a free, open-standard XML-based markup interchange format for standardizing the monitoring and exchange of enzyme kinetic data. It allows scientists to share experimental protocols, results, and kinetic data even if they are using different instruments, electronic laboratory notebooks, or databases.
EnzymeML follows the FAIR Principles (Findable, Accessible, Interoperable, Reusable), ensuring data is easily located, accessed, combined, and reused. It is compatible with the Systems Biology Markup Language, and is supported by the STRENDA Commission. The format continues to be evolved and expanded by an international community.
Development is jointly led by Juergen Pleiss, Frank T. Bergmann, and Santiago Schnell. By promoting data standardization and accessibility, EnzymeML aims to accelerate research in enzyme kinetics and biocatalysis, streamline collaborations, and ultimately contribute to advancements in drug discovery, biotechnology, and our understanding of biological and chemical systems.
Unraveling the Molecular Mechanisms of Rare Disease Mechanisms through Mathematical Modeling
We leverage mathematical modeling and computational approaches to dissect the complex biological mechanisms underlying rare diseases. By studying perturbations in animal models that mirror these diseases, we gain insights into both normal and pathological processes, paving the way for potential therapeutic interventions.
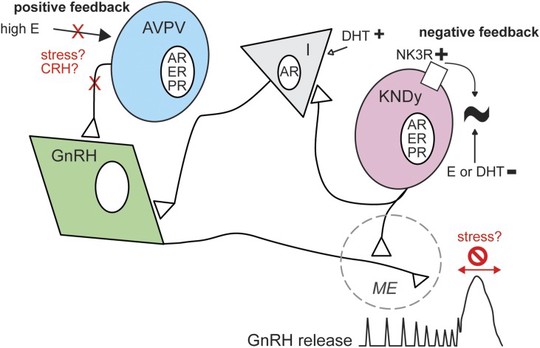
The Schnell Lab is dedicated to understanding the molecular mechanisms of rare diseases, where mathematical modeling and theory aid in identifying the key components leading to disease onset. Our collaborative approach combines experimental biology with mathematical, computational, and statistical methods to model complex interactions of molecules, cells, and tissues in healthy and diseased states. These models act as powerful tools, allowing us to elucidate the molecular mechanisms leading to the onset and progression of disease. By identifying how specific mutations or molecular disruptions lead to disease, we can uncover potential targets for drug development and therapeutic interventions.
Our projects often involve collaborations with both theoretical and experimental researchers. For example, we have developed models that shed light on the hormonal signaling pathways disrupted in Kallmann syndrome, a rare form of infertility. We have also investigated the molecular basis of proinsulin misfolding and its role in cellular toxicity in an idiopathic non-autoimmune type 1 diabetes. Additionally, our models have provided insights into the signaling pathways responsible for impaired intestinal growth in short bowel syndrome, the molecular switches that drive the development of small cell lung cancer, and the unique metabolic features of inflammatory breast cancer.
Animal models with specific genetic alterations that mimic rare diseases serve as a valuable window into disease processes. Studying these models provides us with crucial insights into the normal functioning of biological systems and how these systems break down in disease. Ultimately, our research is driven by the goal of translating our findings into novel treatments and improved outcomes for individuals affected by rare diseases.
You can learn more about current research through our Publications.
The Role of Crowding in Cell Physiology and Biochemistry
The study of macromolecular crowding is indispensable for understanding cellular physiology and biochemistry. We are investigating how crowding affects quantitatively the cellular biochemistry, and qualitatively cellular physiology.
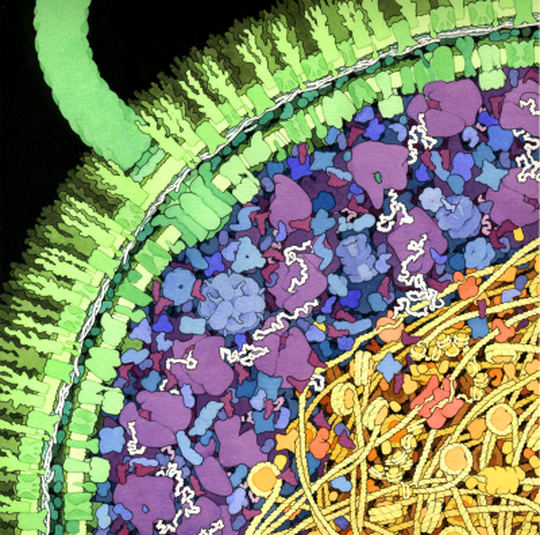
The intracellular environment is densely packed with macromolecules, occupying up to 30% of the cellular volume at concentrations reaching 300 grams per liter. This phenomenon, known as macromolecular crowding, significantly influences cellular biochemistry and physiology.
In these crowded conditions, the rates of biochemical reactions can differ markedly from those in dilute environments. Crowding reduces the diffusion of reactants, slowing diffusion-limited reactions and giving rise to fractal-like reaction kinetics (where reactions exhibit non-classical, complex behaviors). Furthermore, cellular crowded environments are distinct from simple concentrated solutions due to their high molecular diversity. While the overall volume fraction of solute molecules is high, the concentration of any specific species can be relatively low.
Macromolecular crowding has been implicated in various cellular processes, including compartmentalization and phase separation (the formation of distinct liquid-like compartments within cells). Our research group is actively exploring the multifaceted effects of macromolecular crowding on cellular physiology. We are also investigating whether current theories on the origin of cells and life need revision to fully account for this crowded cellular organization.
Measuring and Improving Research University Performance
We are developing new metrics and models to measure and improve performance of scientists, research universities, and academic policies.

Modern research universities are complex ecosystems, requiring a deep understanding of their inner workings to make informed policy and administrative decisions. Scientometrics, the quantitative study of scientific research, and economic modeling provide the tools to achieve this.
Our goal is to develop models that leverage large-scale scientometric and economic data to optimize the use of limited resources, ultimately maximizing research output and societal impact. We investigate the ongoing cost-benefit decisions academic institutions make as they compete for resources and reputation, as well as how scientists respond to incentives and costs related to research funding, scientific quality, and reputation.
By understanding these dynamics, we aim to inform strategies that enhance the efficiency and effectiveness of research universities, foster a thriving academic environment, and ultimately contribute to the advancement of knowledge and innovation.